This week was fun as it was my first experience working in the wet lab. Before I had been strictly doing computational work but this week I was more oriented towards physical lab work. Most of my work this week involved preparing slides for imaging.
I started off by slicing frozen liver tissue using a machine similar to a deli slicer. However it shaved the sample down to 15 micron slices that were fit to be put on a slide for microscope imaging. Here is a picture of my tissue in the “slicer”.
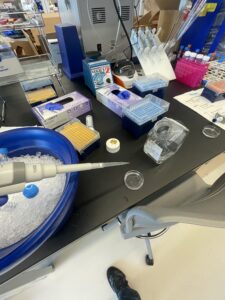
My next step was to fix the tissue with formaldehyde for about 10-20min in order to prevent tissue degradation. The formaldehyde binds to the proteins in the tissue fixing them in place. Another measure I would use to prevent tissue degradation is keeping my sample and equipment cold at all times, either on ice or in the freezer. After fixing the tissue with formaldehyde I would wash the sample with phosphate buffer solution(PBS), not water as PBS has a similar PH and solute concentration to your body, making it more effective. In order to wash the tissue I would pipette PBS onto the sample, let it sit for about 5 minutes, and then suck the liquid back up using another pipette. I would repeat this washing step three times for each sample. In the image below you can see me washing some of my tissue samples, which are in the glass dish on the desk in front of me.
After washing the tissue I would stain it with SiR-DNA, a fluorescent tag that binds to DNA and is used to highlight the nucleus. This stain specifically works well in super-resolution microscopy. After staining the sample for about 10 minutes I would do two more quick washes with PBS. While working with a graduate student, Anthony, we tried to use fiducial beads in a few samples. These are microscopic gold beads that remain in-place when imaging a sample multiple times. It helps in analysis to account for the movement of the tissue during multiple rounds of imaging. However, when he imaged it, it didn’t work that well, probably because it is a new technique in the lab.
After all the work and prep that I just described has been completed, imaging the samples underneath a microscope would be the next step, however I didn’t quite get to that part yet. I did the process I described above a couple times over four days. However, using the microscope I have pictured here,
the samples would look something similar to this. This is an image of a cell stained with DAPI, a tag that also highlights nuclei.
I also did computational stuff this week too. I started a new project in which I am trying to write code for a large set of images. The images are of one sample that went through multiple rounds of imaging causing a shift between some of the images in the x, y, and z dimensions. My code is to find the translation between these images which is turning out to be pretty hard. Running the code is a computationally intense process because it is such a large data set, and the functions I am using take a long time to compute. It takes multiple hours to run my code. I haven’t finished this assignment yet.
Outside of the lab has been fun too. There has been a lot of rain this week so I’ve been outside less but I’ve enjoyed going to the gym, running, cooking food, and playing my guitar. Today (Sunday), I went downtown to see the Cathedral of Saint Paul and Peter and go to mass. It was really cool to check out and it is a very beautiful church. I have it pictured here.




There are no comments published yet.