Welcome to Week 4 of the Computational Biology Internship. My name is Eliram Reyes-Powell and I am interning at University of California, Davis, School of Medicine in the Department of Physiology and Membrane Biology. This is the First Week in the Molecular Dynamics Lab with Dr. Igor Vorobyov, Dr. Slava Bekker, and graduate student John Dawson.
We wrapped up the Molecular Modelling Internship with Dr. Vladimir Yarov-Yarovoy and Mr. Brandon Harris on Monday, June 29th. We went over our results using GALigandDock and compared the original structure to the best structure processed through GALigandDock and the result was excellent and very satisfying.
My best structure compared to the original structure (superposed):
On that same day, Monday, we had the first meeting for the Molecular Dynamics Internship and we went over the essentials of Molecular Dynamics (MD abbreviated). Molecular Dynamics simulations are based on a series of parameters called empirical force fields (which Dr. Slava Bekker talked about in depth). Dr. Vorobyov then explained to us how simulations need supercomputers to run for hours or days to even simulate what a molecule would do in a few nanoseconds or microseconds (for the most powerful computers). It was very interesting to see how complicated it is to simulate such “minute” (relative to us) molecules, since one would expect to see simulations happen rather quickly, but it is quite the contrary. To me, those facts were very overwhelming since that meant I wouldn’t have the chance to do it myself (since it is a very costly and lengthy process). However, I am incredibly grateful that we will get to interpret results from a professional Molecular Dynamics simulation, which I am very excited about because I will be able to use the Calculus and Statistics knowledge I have accumulated so far. (I am also thankful for the great opportunity and for my mentors´ warmth and extensive knowledge they share with us everyday.)
Critical for the internship, Dr. Slava Bekker gave a presentation to us about Valinomycin and all of the incredible feats of which that molecule (a carrier) is capable. We went over Valinomycin’s chemistry, Intermolecular Forces, and behaviors in different solvents. Dr. Bekker told us that Valinomycin’s key feature is that it has the strongest selectivity for potassium ions out of all molecules known to human beings. That selectivity represents an enormous advantage for its use as an antibiotic. Impressively, it is being studied for its use as an antiviral for COVID-19 by many scientists.
We were then encouraged to use the free (since supercomputers can cost millions of dollars) and fast (a few hours compared to what a professional simulation takes) extension of the program Visual Molecular Dynamics (VMD), QwikMD (Quick Molecular Dynamics), using Nanoscale Molecular Dynamics (NAMD) to visualize and experience what a simulation is like (very motivating and exciting).
I was able to use VMD to load some of the structures I had been working with in the past weeks to observe them in a “dynamic” environment. I visualized the different forms valinomycin can take in order to understand the molecule better.
Next, Mr. John Dawson showed us all of the features of VMD we need to know about for the internship and he talked about some of his favorite applications of the program and Molecular Dynamics in general. It was very challenging to follow along all the steps Mr. Dawson performed live, but I managed to replicate his actions and I was able to run his trajectory for the bracelet-form of valinomycin that had about 3 nanoseconds of simulation.
With that knowledge, I decided to obtain my own trajectory from a simulation.
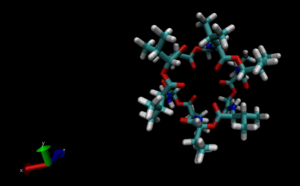
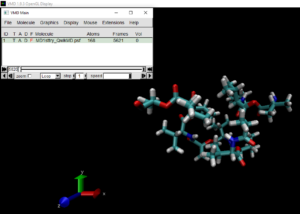
I, fortunately, had run a simulation with QwikMD for about 22 hours a day before. I needed my mentors´ guidance to obtain the trajectory and learn where all of the output files need to go, so it was challenging (but very exciting). I obtained a great trajectory and data of how Valinomycin changed under certain controlled conditions in an implicit solvent for 9.952 ns (Very close to 10 ns!! ).
Valinomycin before (Left) and after (Right) the 22 hour simulation :
It has been great to learn about Molecular Dynamics and how this field complements Molecular Modelling very well. In that sense, all of the knowledge I am gaining now can be used for when I go back to work with Mr. Harris, because I can have a better understanding of the molecular world. I have learnt a lot and the complexity of the Molecular Dynamics field makes me want to learn more about it everyday, even if simulations take a while. I am willing to wait, it is definitely worth it when you see the results.
When not in my internship, I spent most of my time preparing for my SAT or reading a book I got a few days ago: “Six easy pieces”, by Richard Feynman. I visited my grandparents and spent some time helping them in their ranch.

There are no comments published yet.