Hello everyone, I’m going to be telling you about my wonderful fourth week learning about computational biology at CMU.
This week marked the start of a new portion of my internship. For the first three weeks I was primarily in lectures learning the fundamentals of the field; however, now the process is far different. The remainder of my time at this internship is being spent following around the knowledgable Joshua Kangas and doing the tasks he gives me. He and I are doing much more focused work, and assisting him in his various endeavors has been truly eye opening. The vast majority of what I’ve been working on can be split into two sections, so I’ll briefly speak about each of those here.
Firstly, I’ll talk about BioActive. This is the project that I have the least personal involvement in, since it was ongoing before I arrived, but it is still highly interesting. Several undergraduate students along with a PhD student (whose work is slightly separate from the rest) under Josh are working to create software that will allow for active machine learning programs to determine future experiments in a campaign. What all that really means is that they want to create a program that, given a search space (you can view this as a huge grid, for example, two drugs where we want to know which ratio of the two will provide the best effect, the x axis being quantities of Drug A and the y axis being quantities of drug B), will try to develop an idea of what that search space looks like without actually doing all of the necessary experiments (it might do a few experiments and then learn that too much of Drug A is bad so it will prioritize trying smaller amounts of Drug A in future experiments). If all of this sounds complicated, that’s probably because it is and even after a week of hearing about all of this I don’t fully understand the mechanics behind how their algorithm selects which experiments to try next. Regardless, it’s an extremely interesting topic and one that I am excited to learn more about.
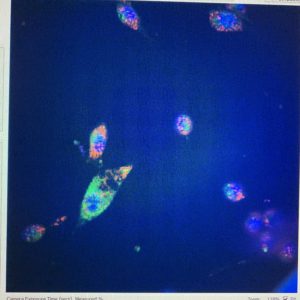
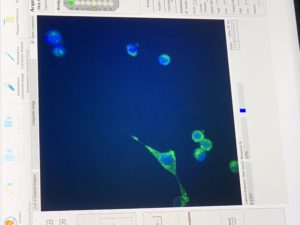
Next, I’ll speak about what I’ve been spending most of my time on: machine automation in the lab. This coming semester Josh is going to be teaching a graduate course on machine automation. This is all about using machines to do lab work for you. Since they can usually do it more quickly and more precisely than you, you can acquire more, and more accurate data. Soon CMU will have a room that does biological experiments completely without human interaction. You will simply load the machines with your samples and then give them the “recipe” for what you want to do. Then a robot arm will load your samples into different machines and those machines will automatically complete your task. The room isn’t built yet, but some of the fancy machines have already arrived and I’m getting to play with some of them. This includes a liquids handler (which moves around liquids by pipetting them, a critical process in most biological experiments) and the CX7 microscope which I talked about in my last Blog. Both are extremely cool and I feel very privileged to be allowed to use them. I’ll try to attach a video of the liquid handler executing some of my commands as well as some photos that the CX7 gave me of cells with florescent dyes added to mark the mitochondria, DNA and lysosomes.
Outside of work I have mostly been catching up on some reading, though I did get some friends together to go see John Wick 3, which was pretty fun. We’ve also taken to playing Jackbox and other party games in one of the lounges of our dorm, which has been great.
I can’t wait to tell you all about my continued adventures next week and I hope that everyone is having a great summer so far!
The liquid handler: IMG_2163

FASCINATING STUFF JOSH!!! Love reading your weekly updates!!!!